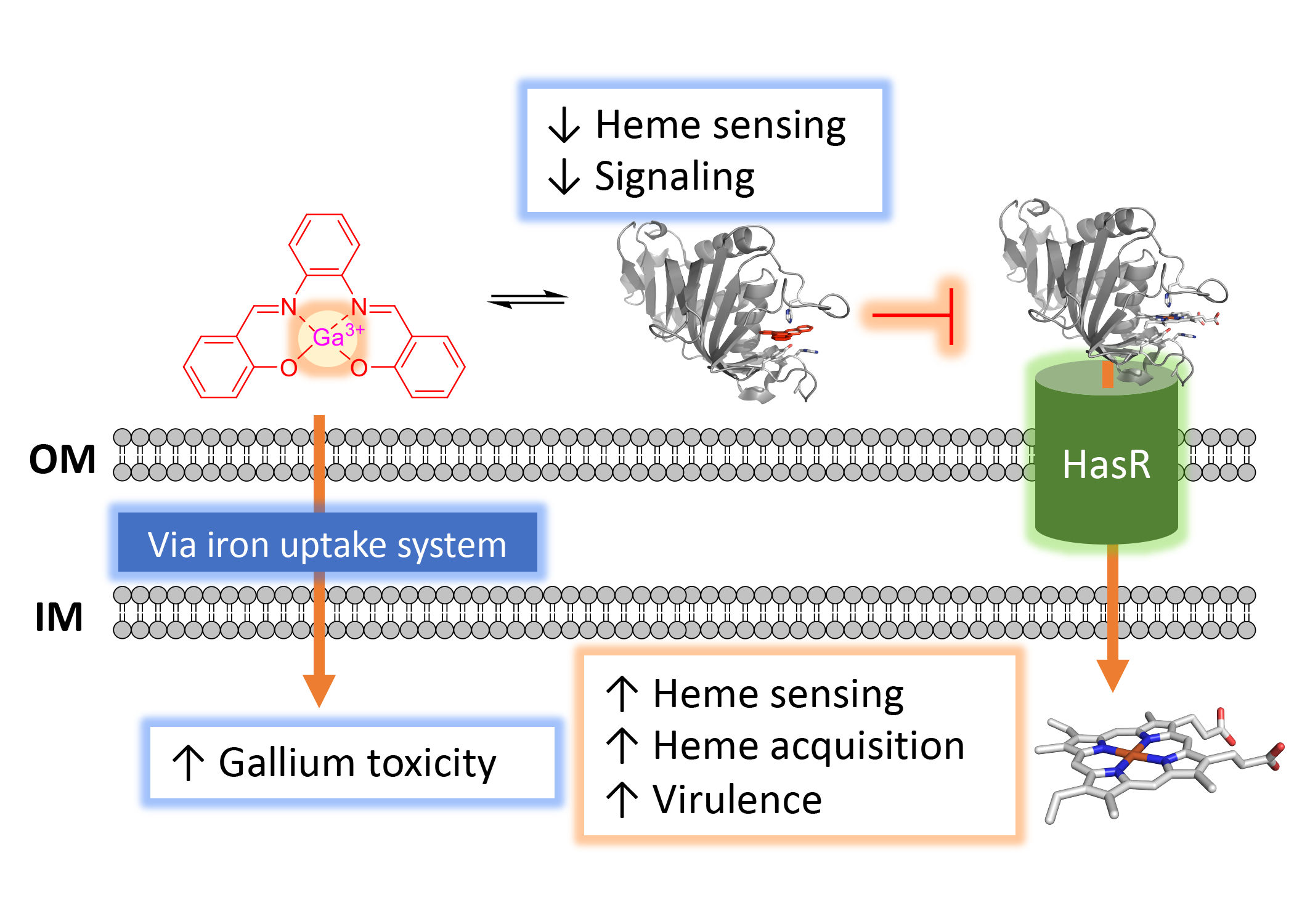
Iron is an essential micronutrient for all living organisms and in pathogenic bacteria it is intricately linked to their virulence. During infection, iron within the host is scarce and bacteria must employ a complex array of iron uptake and regulatory pathways to maintain iron homeostasis and promote virulence. The focus of the Wilks laboratory is to understand how iron acquisition, and in particular heme, contribute to bacterial pathogenesis. Heme represents an abundant source of iron within the host, and pathogens have evolved sophisticated mechanisms by which they acquire and utilize heme. We are particularly interested in the contributions of heme acquisition to the virulence of P. aeruginosa, an opportunistic pathogen of high incidence in hospital acquired infections and in cystic fibrosis patients. Utilizing a multi-disciplinary approach that includes bacterial genetics, proteomics and metabolomics, as well as biochemical and biophysical approaches, we are determining the mechanisms by which P. aeruginosa heme acquisition contributes to iron-homeostasis and pathogenesis. These studies have led to the rational design of novel therapeutic strategies that reduce virulence through global effects on metabolism.
Structure-Function and Regulation of the P. aeruginosa Heme Uptake Systems
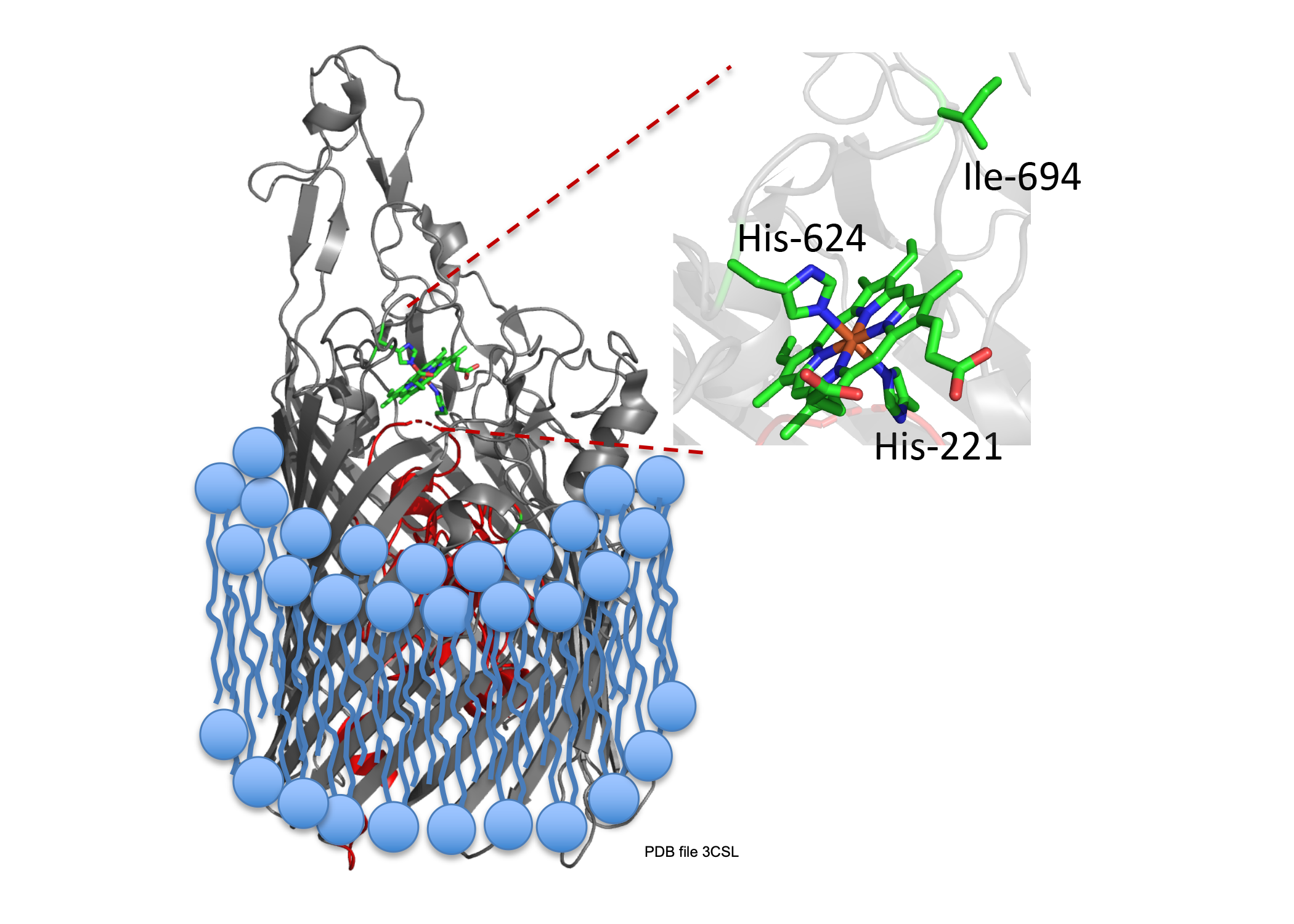
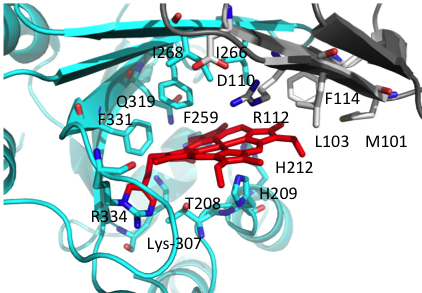
A long term goal of my research has been to understand the contributions of the heme assimilation (Has) and Pseudomonas heme utilization (Phu) systems to heme acquisition and pathogenesis.The HasR and PhuR outer-membrane receptors have distinct coordination motifs that contribute to their respective functions in heme sensing and transport, respectively. The hemophore HasAp and receptor HasR are transcriptionally regulated by the extra cytoplasmic function sigma factor, HasI and post-transcriptionally regulated by the product of heme metabolism, biliverdin IX beta. Metabolite dependent post-transcriptional regulation is a rapid and energetically cost effective means of amplifying the extracellular HasAp-HasR signal. The metabolite dependent post-transcriptional regulation of the Has signaling cascade represents a new paradigm in iron-starvation ECF factor dependent cell signaling. We are further investigating the biliverdin-dependent post-transcriptional regulation of heme uptake. In contrast the Phu system, in addition to the outer membrane receptor PhuR, encodes the cytoplasmic ABC-transporter PhuUV and the intracellular cytoplasmic heme chaperone, PhuS. This system is the high capacity heme transport system, which is partly due to the unique His-Tyr ligand heme coordination of PhuR, an emerging motif in high affinity heme transport proteins. Both the Has and Phu systems have been shown to have distinct roles in adaptation and infection within the host. We are currently working on 1) Heme and iron regulation of the Has and Phu systems and how their complex interplay contributes to virulence and pathogenesis and 2) Determining the molecular mechanisms of heme signaling and transport by the Has and Phu systems. We predict such studies will identify potential therapeutic strategies targeting heme metabolism.
Intracellular Heme Trafficking and Homeostasis
We have recently characterized the P. aeruginosa cytoplasmic heme binding binding protein PhuS as a specific heme chaperone to the iron-regulated heme oxygenase (HemO). Biophysical studies have revealed heme coordination drives a conformational “induced fit” required for interaction with HemO. Furthermore, we describe a novel mechanism of heme transfer for the cytoplasmic heme binding proteins, where a ligand induced conformational change facilitates protein-protein interaction, and the free energy derived from this interaction triggers heme transfer via a histidine relay. Isotopic labeling (13C-heme) and transcriptional analysis further confirmed that HemO drives the metabolic flux of heme into the cell with PhuS acting as a “control-valve”. Coupling the metabolic heme flux through HemO to the regulatory networks provides a novel mechanism for P. aeruginosa to rapidly respond and adapt to sudden changes in heme and iron homeostasis. These findings provide a platform for future studies directed at understanding the central role heme metabolism in the virulence and pathogenesis of P. aeruginosa.
Antimicrobial Inhibitor Development
Pseudomonas aeruginosa is an opportunistic human pathogen that causes severe infections in individuals with underlying conditions, including cystic fibrosis (CF). P. aeruginosa requires iron for virulence, biofilm formation, and is resistant to many antibiotics. We hypothesize that heme is a significant source of iron during infection and that inhibition of heme sensing and utilization may lead to novel therapeutic strategies. We have three ongoing projects targeting a) the Has heme sensing system with a series of heme mimetics; b) HemO via small molecule inhibitors directed toward the active site as well as an allosteric site and c) A novel biliverdin-dependent riboswitch. We utilize variety of biochemical and biophysical approaches including high-throughput screening and fragment-based NMR and HDX-MS. Such novel approaches may lead to the development of novel antimicrobials that reduce the pressure to undergo mutation that lead to resistance.